A radar or spider-web plot is a great way to visualise multivariate data when there are three or more continuous variables and several groups (categorical data). In this example data are first prepared and then two methods to create a radar plot will be described.
Preparation of the data
Download and open the tests.rda dataset for this example and open it in R. The data set contains the results of 5 tests as a classifier for infection. The gold standard for infection is indicated by the variable ‘standard’. All variables are binary (0, 1) and data that are not available are indicated by ‘NA’. The data can be shown by:
tests
test1 test2 test3 test4 test5 standard
1 0 1 0 0 0 0
2 1 NA NA NA 0 0
.....
.....
55 1 0 1 0 0 0It is straightforward to create a confusion table that compare the classifier to the gold standard:
test1 <- table(tests$test1, tests$standard)
test2 <- table(tests$test2, tests$standard)
test3 <- table(tests$test3, tests$standard)
test4 <- table(tests$test4, tests$standard)
test5 <- table(tests$test5, tests$standard)The show the confusion table for test 3:
test3
0 1
0 35 2
1 10 4The positive predictive value, negative predictive value, sensitivity, specificity and accuracy can be found using the epiR package1. Unfortunately, the data in the confusion table is in the wrong order to be inserted into the epi.tests function of the epiR package. To change the order of the matrix:
matrix_test1 <-matrix(c(test1[2,2],test1[1,2],test1[2,1],test1[1,1]), ncol = 2)
matrix_test2 <-matrix(c(test2[2,2],test2[1,2],test2[2,1],test2[1,1]), ncol = 2)
matrix_test3 <-matrix(c(test3[2,2],test3[1,2],test3[2,1],test3[1,1]), ncol = 2)
matrix_test4 <-matrix(c(test4[2,2],test4[1,2],test4[2,1],test4[1,1]), ncol = 2)
matrix_test5 <-matrix(c(test5[2,2],test5[1,2],test5[2,1],test5[1,1]), ncol = 2)To show, for example, the matrix of test 3:
matrix_test3
[,1] [,2]
[1,] 4 10
[2,] 2 35To summarise the positive predictive value, negative predictive value, sensitivity, specificity and accuracy using the epiR package2, one large matrix (called overall) can be build after the epiR package has been loaded:
library(epiR)
overall <- matrix(c(
epi.tests(matrix_test1)$detail$est[epi.tests(matrix_test1)$detail$statistic == "pv.pos"],
epi.tests(matrix_test1)$detail$est[epi.tests(matrix_test1)$detail$statistic == "pv.neg"],
epi.tests(matrix_test1)$detail$est[epi.tests(matrix_test1)$detail$statistic == "se"],
epi.tests(matrix_test1)$detail$est[epi.tests(matrix_test1)$detail$statistic == "sp"],
epi.tests(matrix_test1)$detail$est[epi.tests(matrix_test1)$detail$statistic == "diag.ac"],
epi.tests(matrix_test2)$detail$est[epi.tests(matrix_test2)$detail$statistic == "pv.pos"],
epi.tests(matrix_test2)$detail$est[epi.tests(matrix_test2)$detail$statistic == "pv.neg"],
epi.tests(matrix_test2)$detail$est[epi.tests(matrix_test2)$detail$statistic == "se"],
epi.tests(matrix_test2)$detail$est[epi.tests(matrix_test2)$detail$statistic == "sp"],
epi.tests(matrix_test2)$detail$est[epi.tests(matrix_test2)$detail$statistic == "diag.ac"],
epi.tests(matrix_test3)$detail$est[epi.tests(matrix_test3)$detail$statistic == "pv.pos"],
epi.tests(matrix_test3)$detail$est[epi.tests(matrix_test3)$detail$statistic == "pv.neg"],
epi.tests(matrix_test3)$detail$est[epi.tests(matrix_test3)$detail$statistic == "se"],
epi.tests(matrix_test3)$detail$est[epi.tests(matrix_test3)$detail$statistic == "sp"],
epi.tests(matrix_test3)$detail$est[epi.tests(matrix_test3)$detail$statistic == "diag.ac"],
epi.tests(matrix_test4)$detail$est[epi.tests(matrix_test4)$detail$statistic == "pv.pos"],
epi.tests(matrix_test4)$detail$est[epi.tests(matrix_test4)$detail$statistic == "pv.neg"],
epi.tests(matrix_test4)$detail$est[epi.tests(matrix_test4)$detail$statistic == "se"],
epi.tests(matrix_test4)$detail$est[epi.tests(matrix_test4)$detail$statistic == "sp"],
epi.tests(matrix_test4)$detail$est[epi.tests(matrix_test4)$detail$statistic == "diag.ac"],
epi.tests(matrix_test5)$detail$est[epi.tests(matrix_test5)$detail$statistic == "pv.pos"],
epi.tests(matrix_test5)$detail$est[epi.tests(matrix_test5)$detail$statistic == "pv.neg"],
epi.tests(matrix_test5)$detail$est[epi.tests(matrix_test5)$detail$statistic == "se"],
epi.tests(matrix_test5)$detail$est[epi.tests(matrix_test5)$detail$statistic == "sp"],
epi.tests(matrix_test5)$detail$est[epi.tests(matrix_test5)$detail$statistic == "diag.ac"]),
ncol = 5)To show the matrix, just call the object:
overall
[,1] [,2] [,3] [,4] [,5]
[1,] 0.1724138 0.2500000 0.2857143 0.6000000 0.8000000
[2,] 0.9615385 0.9736842 0.9459459 0.9772727 0.9795918
[3,] 0.8333333 0.7500000 0.6666667 0.7500000 0.8000000
[4,] 0.5102041 0.8043478 0.7777778 0.9555556 0.9795918
[5,] 0.5454545 0.8000000 0.7647059 0.9387755 0.9629630Apply collumn and row names and round the values:
colnames(overall) <- c('Test1','Test2','Test3','Test4','Test5')
rownames(overall) <- c('PPV', 'NPV', 'Sens', 'Spec', 'Acc')
round(overall*100,0)
Test1 Test2 Test3 Test4 Test5
PPV 17 25 29 60 80
NPV 96 97 95 98 98
Sens 83 75 67 75 80
Spec 51 80 78 96 98
Acc 55 80 76 94 96The matrix can be transposed (t) and converted to a data frame, called radar, for subsequent analysis:
radar <- t(round(overall*100,0))
radar <- as.data.frame(radar)
radar
PPV NPV Sens Spec Acc
Test1 17 96 83 51 55
Test2 25 97 75 80 80
Test3 29 95 67 78 76
Test4 60 98 75 96 94
Test5 80 98 80 98 96Two Methods to create a radar plot will be described:
- Using the fmsb package3(easier, but less versatile)
- Using function as provided by LePennec4 using the ggplot2 package5
Method 1:
Make sure the fmsb6 package is loaded and create a simple radar plot:
library(fmsb)
radarchart(radar, maxmin = F)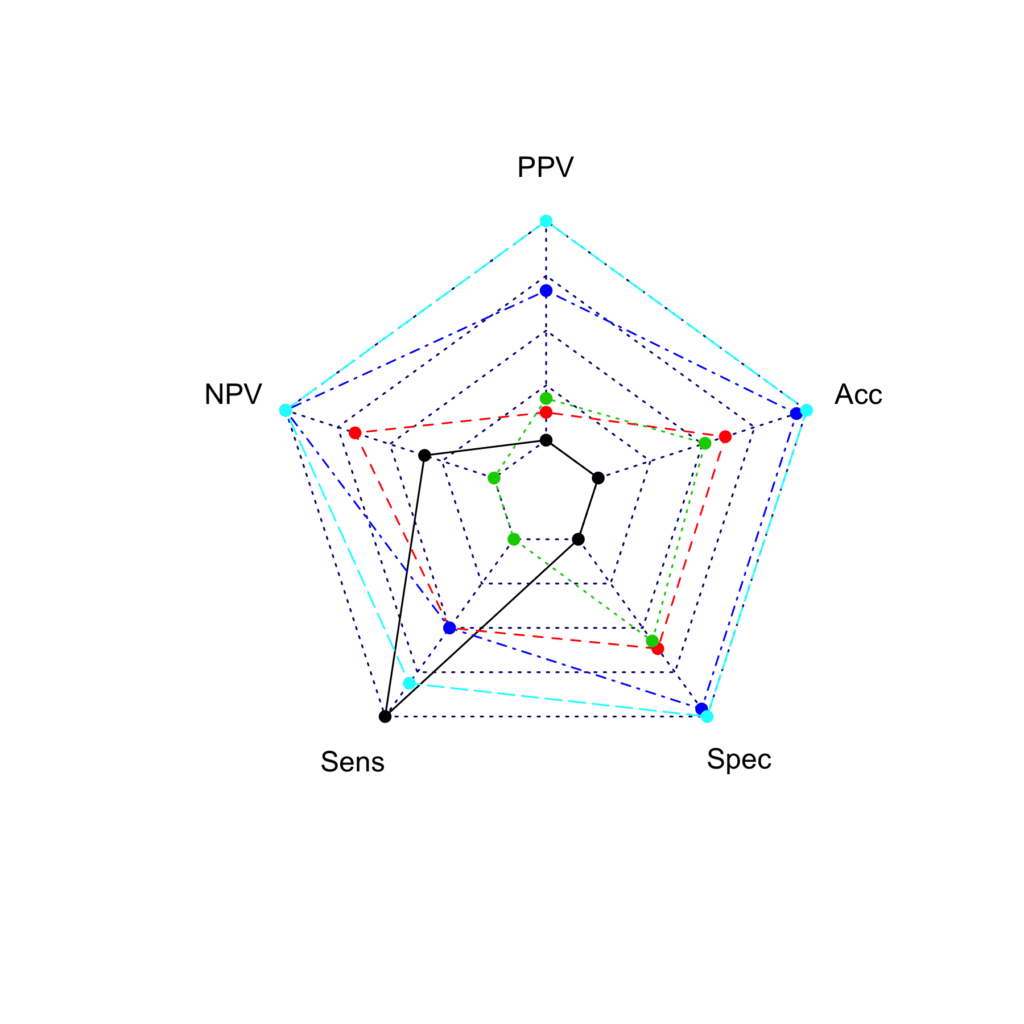
The add maximum (100) and minimum (0) values to the plot, they need to be inserted to the first and second row of the data frame respectively:
radar2 <- rbind(rep(100,5) , rep(0,5) , radar)
radarchart(radar2)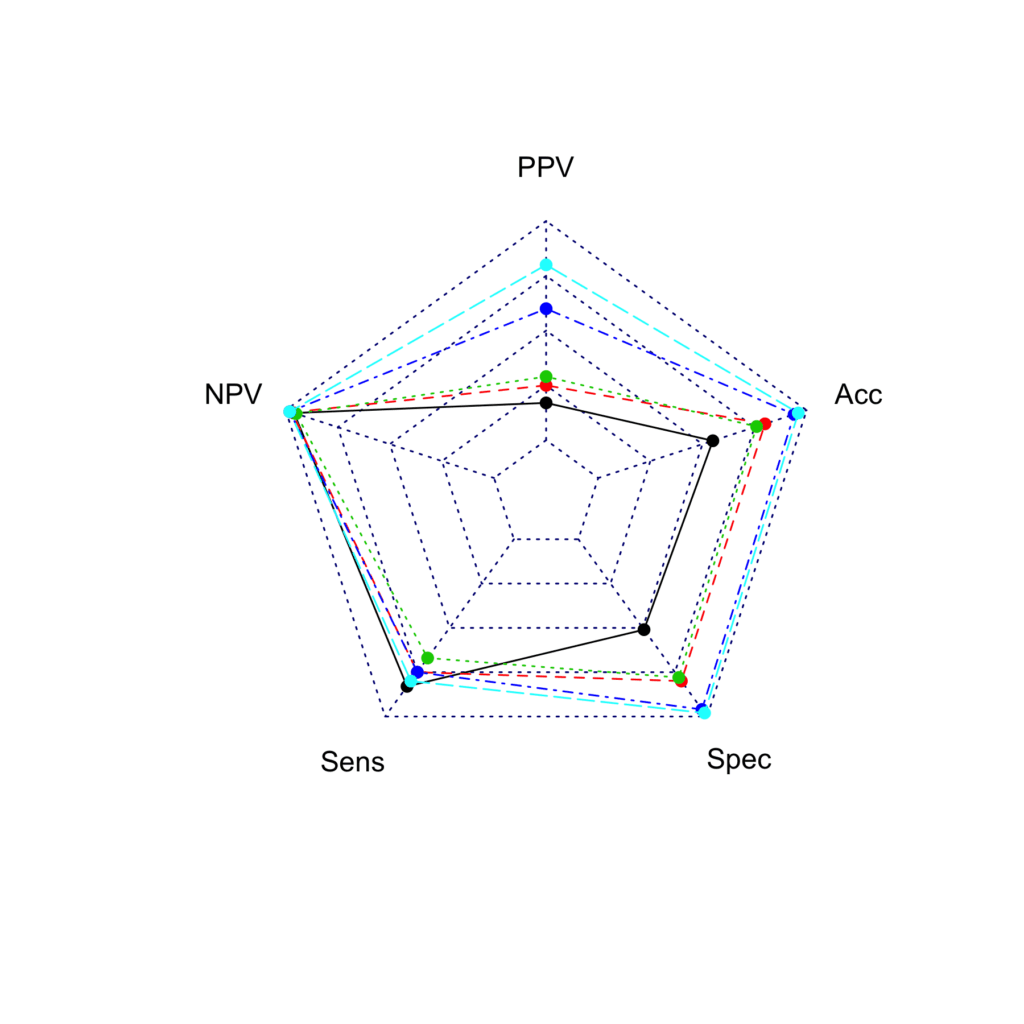
A better looking plot can be obtained by setting the arguments as per the manual of the package:
colours <- c( 'tomato','orchid','grey','gold', 'seagreen')
radarchart(radar, axistype=1 , maxmin=F, pcol=colours, plwd=4, plty=1,
cglcol="grey", cglty=1, axislabcol="black", cglwd=0.8, vlcex=0.8)
legend(x=0.7, y=1.2, legend = rownames(radar), bty = "n", pch=20, col=colours, text.col = "grey", cex=1.2, pt.cex=3)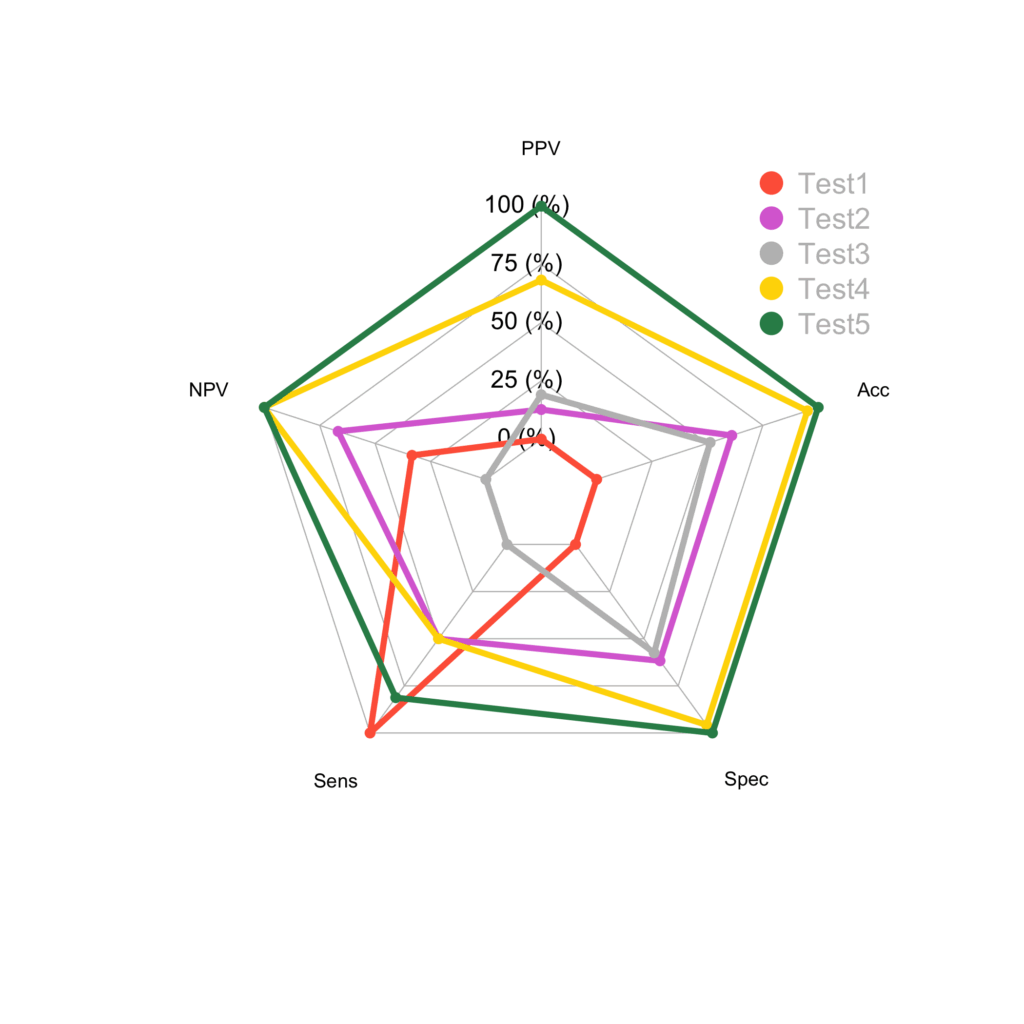
Method 2:
This method is more versatile, but requires more packages to be loaded. Plots are created using the ggplot2 package7 and a function from LePennec8.
Make sure the required packages , including reshape29, are loaded:
library(reshape2)
library(ggplot2)Have a look at the data frame again:
radar
PPV NPV Sens Spec Acc
Test1 17 96 83 51 55
Test2 25 97 75 80 80
Test3 29 95 67 78 76
Test4 60 98 75 96 94
Test5 80 98 80 98 96The data frame needs to contain a varaible that contains the test; therefore define a new data frame and call it radar3:
test <- c("Test 1", "Test 2", "Test 3", "Test 4", "Test 5")
radar3 <- cbind(test, radar)
radar3
test PPV NPV Sens Spec Acc
Test1 Test 1 17 96 83 51 55
Test2 Test 2 25 97 75 80 80
Test3 Test 3 29 95 67 78 76
Test4 Test 4 60 98 75 96 94
Test5 Test 5 80 98 80 98 96Now melt the data using the reshape2 package10:
radar4 <- melt(radar3)
radar4
test variable value
1 Test 1 PPV 17
2 Test 2 PPV 25
.....
.....
25 Test 5 Acc 96and give collumn names:
colnames(radar4) <- c('Test', 'Descript', 'Value')
radar4
Test Descript Value
1 Test 1 PPV 17
2 Test 2 PPV 25
.....
.....
25 Test 5 Acc 96LePennec11 has defined a function that creates radar charts with the ggplot2 package. Load the function:
coord_radar <- function (theta = c("x", "y"), start = 0, direction = 1) {
theta <- match.arg(theta)
r <- if (theta == "x") "y" else "x"
ggproto(
"CoordRadar", CoordPolar,
theta = theta,
r = r,
start = start,
direction = sign(direction),
is_linear = function(coord) TRUE
)
}Finally, build the plot using the ggplot2 package:
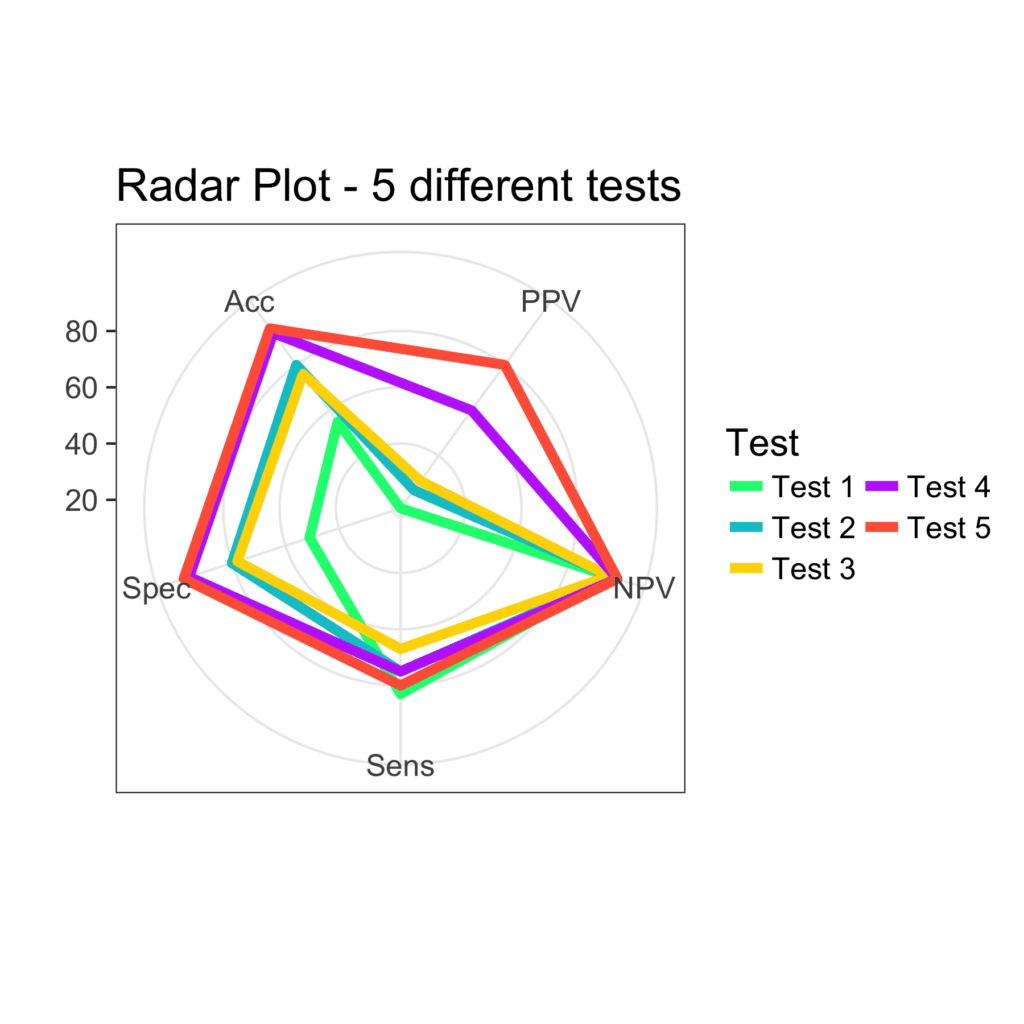
ggplot(radar4, aes(x = Descript, y = Value)) +
geom_polygon(aes(group = Test, color = Test), fill = NA, size = 2, show.legend = FALSE) +
geom_line(aes(group = Test, color = Test), size = 2) +
xlab("") + ylab("") +
guides(color = guide_legend(ncol=2)) +
scale_color_manual(values = c('springgreen', 'turquoise3', 'gold', 'darkorchid1', 'tomato')) +
coord_radar() +
ggtitle('Radar Plot - 5 different tests') +
theme_bw(base_size = 16)