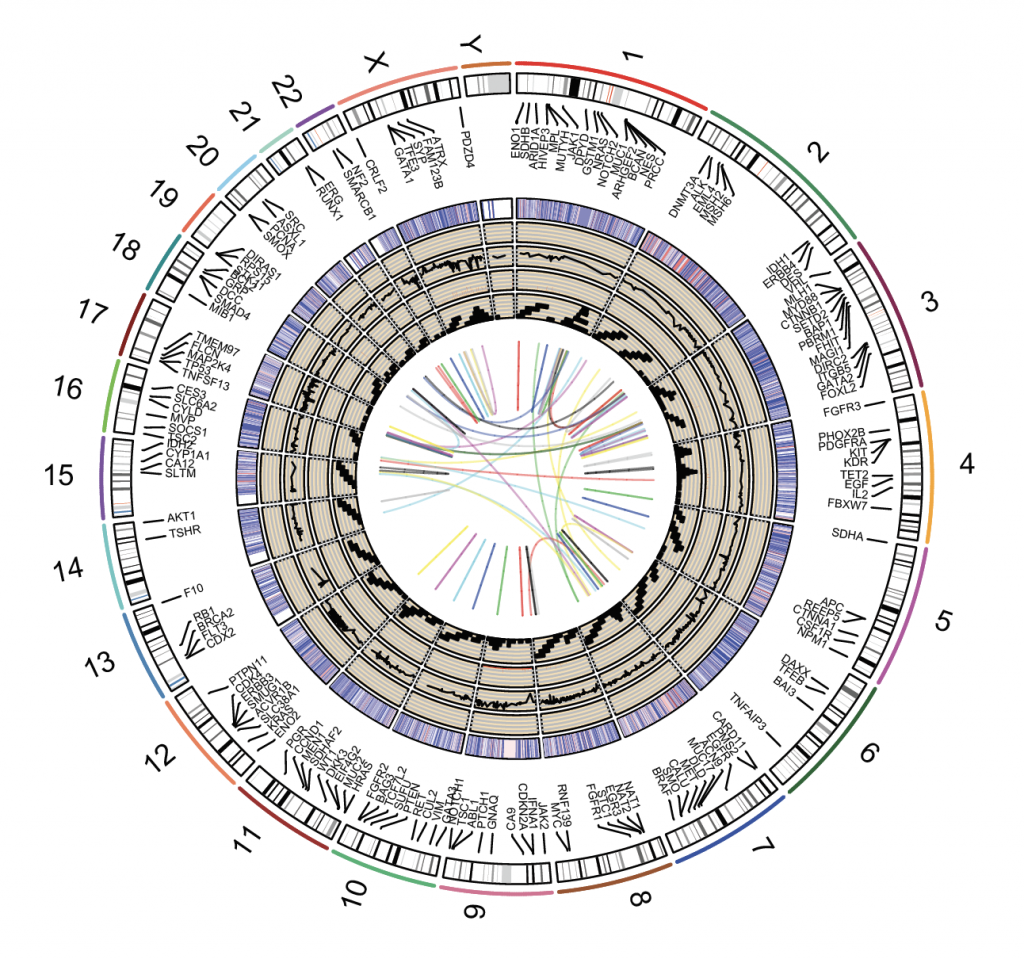
RCircos 1 can be used to make chromosome ideograms. The example below uses the hg19 assembly (GRCh37) as reference. However, the newer (hg38 / GRCh38) can also be downloaded, manipulated and used instead. It is important to use the appropriate reference assembly when working with genomic data.
Open the package and create a default plot using the build in data with different tracks as example (data are printed out for clarity):
# RCircos, to make circos plots for genomic data
library(RCircos)
data(UCSC.HG19.Human.CytoBandIdeogram)
cyto.info <- UCSC.HG19.Human.CytoBandIdeogram
head(cyto.info)
Chromosome ChromStart ChromEnd Band Stain
1 chr1 0 2300000 p36.33 gneg
2 chr1 2300000 5400000 p36.32 gpos25
3 chr1 5400000 7200000 p36.31 gneg
4 chr1 7200000 9200000 p36.23 gpos25
5 chr1 9200000 12700000 p36.22 gneg
6 chr1 12700000 16200000 p36.21 gpos50
RCircos.Set.Core.Components(cyto.info, chr.exclude = NULL, tracks.inside = 10, tracks.outside = 0)
RCircos.Core.Components initialized.
Type ?RCircos.Reset.Plot.Parameters to see how to modify the core components.
# initiallize graphic devide
dev.new()
# set plot area with default values:
RCircos.Set.Plot.Area()
# plot the chromosome ideogram:
RCircos.Chromosome.Ideogram.Plot()
# plot the connectors and gene names on the 1st and 2nd track
# gene names are held in the 4th column
data(RCircos.Gene.Label.Data)
head(RCircos.Gene.Label.Data)
Chromosome chromStart chromEnd Gene
1 chr1 8921418 8934967 ENO1
2 chr1 17345375 17380514 SDHB
3 chr1 27022894 27107247 ARID1A
4 chr1 41976121 42501596 HIVEP3
5 chr1 43803519 43818443 MPL
6 chr1 45794977 45805926 MUTYH
RCircos.Gene.Connector.Plot(RCircos.Gene.Label.Data, track.num = 1, side = “in”)
Not all labels will be plotted.
Type RCircos.Get.Gene.Name.Plot.Parameters()
to see the number of labels for each chromosome.
RCircos.Gene.Name.Plot(RCircos.Gene.Label.Data, name.col = 4, track.num = 2, side = “in”)
Not all labels will be plotted.
Type RCircos.Get.Gene.Name.Plot.Parameters()
to see the number of labels for each chromosome.
# heatmap plot on the 5th track and data values on the sixth
data(RCircos.Heatmap.Data)
head(RCircos.Heatmap.Data)
Chromosome chromStart chromEnd GeneName X786.O A498 A549.ATCC ACHN BT.549 CAKI.1
1 chr1 934341 935552 HES4 6.75781 7.38773 6.47890 6.05517 8.85062 7.00307
2 chr1 948846 949919 ISG15 7.56297 10.49590 5.89893 7.58095 12.08470 7.81459
3 chr1 1138887 1142089 TNFRSF18 4.69775 4.55593 4.38970 4.50064 4.47525 4.47721
4 chr1 1270657 1284492 DVL1 7.76886 7.52194 6.87125 7.03517 7.65386 7.69733
5 chr1 1288070 1293915 MXRA8 4.49805 4.72032 4.62207 4.58575 5.66389 4.93499
6 chr1 1592938 1624243 SLC35E2B 8.73104 8.10229 8.36599 9.04116 9.24175 9.89727
RCircos.Heatmap.Plot(RCircos.Heatmap.Data, data.col = 6, track.num = 5, side = “in”)
# scatterplot on the 6th track with data values on the fifth
# colours will be red for values >= 1 and blue for <= -1
data(RCircos.Scatter.Data)
head(RCircos.Scatter.Data)
chromosome start stop num.mark seg.mean
1 1 61735 228706 18 -0.4459
2 1 228729 356443 10 0.5624
3 1 356542 564621 4 -0.9035
4 1 603590 1704138 227 0.3545
5 1 1709023 1711414 6 1.2565
6 1 1714558 12862252 6276 0.4027
# some data not in ideogram: error
# this is because the chromosome field has only the number, rather than chr1; so just change the file and put chr in front of each chromosome number in the first column!
RCircos.Scatter.Data$chromosome <- paste(“chr”, RCircos.Scatter.Data$chromosome, sep = “”)
head(RCircos.Scatter.Data)
chromosome start stop num.mark seg.mean
1 chr1 61735 228706 18 -0.4459
2 chr1 228729 356443 10 0.5624
3 chr1 356542 564621 4 -0.9035
4 chr1 603590 1704138 227 0.3545
5 chr1 1709023 1711414 6 1.2565
6 chr1 1714558 12862252 6276 0.4027
RCircos.Scatter.Plot(RCircos.Scatter.Data, data.col = 5, track.num = 6, side = “in”)
# line plot on the 7th track with data values on the fifth
data(RCircos.Line.Data)
head(RCircos.Line.Data)
chromosome start stop num.mark seg.mean
1 1 61735 16895627 8732 0.1797
2 1 16896821 17212714 105 -0.2117
3 1 17214822 25574471 5321 0.1751
4 1 25574707 25662212 37 0.5064
5 1 25663310 30741496 2400 0.1384
6 1 30741656 30745210 3 -1.4742
# some data not in ideogram: error
# this is because the chromosome field has only the number, rather than chr1 (as above).
RCircos.Line.Data$chromosome <- paste(“chr”, RCircos.Line.Data$chromosome, sep = “”)
head(RCircos.Line.Data)
chromosome start stop num.mark seg.mean
1 chr1 61735 16895627 8732 0.1797
2 chr1 16896821 17212714 105 -0.2117
3 chr1 17214822 25574471 5321 0.1751
4 chr1 25574707 25662212 37 0.5064
5 chr1 25663310 30741496 2400 0.1384
6 chr1 30741656 30745210 3 -1.4742
RCircos.Line.Plot(RCircos.Line.Data, data.col = 5, track.num = 7, side = “in”)
# histogram on the 8th track with data values in the fourth column
data(RCircos.Histogram.Data)
head(RCircos.Histogram.Data)
Chromosome chromStart chromEnd Data
1 chr1 45000000 49999999 0.070859
2 chr1 55000000 59999999 0.300460
3 chr1 60000000 64999999 0.125421
4 chr1 70000000 74999999 0.158156
5 chr1 75000000 79999999 0.163540
6 chr1 80000000 84999999 0.342921
RCircos.Histogram.Plot(RCircos.Histogram.Data, data.col = 4, track.num = 8, side = “in”)
# tile plot on the 9th track
data(RCircos.Tile.Data)
head(RCircos.Tile.Data)
Chromosome chromStart chromEnd
1 chr1 0 23900000
2 chr1 12700000 44100000
3 chr1 28000000 68900000
4 chr1 59000000 94700000
5 chr1 99700000 120600000
6 chr1 147000000 234700000
RCircos.Tile.Plot(RCircos.Tile.Data, track.num = 9, side = “in”)
Tiles plot may use more than one track. Please select correct area for next track if necessary.
# Link line plot on the 9th track
data(RCircos.Link.Data)
head(RCircos.Link.Data)
Chromosome chromStart chromEnd Chromosome.1 chromStart.1 chromEnd.1
1 chr1 8284703 8285399 chr1 8285752 8286389
2 chr1 85980143 85980624 chr7 123161313 123161687
3 chr1 118069850 118070319 chr1 118070329 118070689
4 chr1 167077258 167077658 chr1 169764630 169764965
5 chr1 171671272 171671550 chr1 179790879 179791292
6 chr1 174333479 174333875 chr6 101861516 101861840
RCircos.Link.Plot(RCircos.Link.Data, track.num = 11, by.chromosome = FALSE)
Depending on the speed of your computer, it may take some time for the plot to be draw: